x <- tryCatch( "OK",
warning=function(w){
return(paste( "Warning:", conditionMessage(w)));
},
error = function(e) {
return(paste( "Error:", conditionMessage(e)));
},
finally={
print("This is try-catch test. check the output.")
});
print(x);
x <- tryCatch( warning("got a warning!"),
warning=function(w){
return(paste( "Warning:", conditionMessage(w)));
},
error = function(e) {
return(paste( "Error:", conditionMessage(e)));
},
finally={
print("This is try-catch test. check the output.")
});
print(x);
x <- tryCatch( stop("an error occured!"),
warning=function(w){
return(paste( "Warning:", conditionMessage(w)));
},
error = function(e) {
return(paste( "Error:", conditionMessage(e)));
},
finally={
print("This is try-catch test. check the output.")
});
print(x);
December 30, 2013
Simple try-catch code in R
Here is a simple code sample to demonstrate tryCatch function in R.
December 29, 2013
How to change the limit of uploaded file size in php?
Edit the following configurations in the php.ini file, and then restart webserver (apache).
You may find the php.ini file in /etc/php5/apache2 directory of ubuntu. If you forget the location of ini file, you may find it by running a simple php file as below.
upload_max_filesize = 10M post_max_size = 20M
You may find the php.ini file in /etc/php5/apache2 directory of ubuntu. If you forget the location of ini file, you may find it by running a simple php file as below.
<?php phpinfo(); ?>
Note: you will probably require write permission to edit the php.ini file.
For more information, you may visit this page.
December 28, 2013
Automatic mapping (or conversion) among different biological databases
biomaRt is an R package to map among different biological databases. Here is a simple code-snippet to convert human gene id to gene symbol.
library(biomaRt)
ensembl = useMart("ensembl",dataset="hsapiens_gene_ensembl")
symbols = getBM(attributes=c('entrezgene','hgnc_symbol'), filters='entrezgene', values=c("7157","5601"), mart=ensembl)
Installation note:
- In Ubuntu, you may need to install 'libxml2-dev' and 'libcurl4-openssl-dev'.
November 15, 2013
String Reverse in R
String reversing is a common operation especially in Bioinformatics. However, this function is not provided in R, not even in the "stringr" package. Here you can find the code:
str_reverse <- function(x){
return(sapply(lapply(strsplit(x, NULL), rev), paste, collapse=""))
}
s <- "abc"
s1 <- str_reverse(s)
print(s1)
November 4, 2013
Code Highlighing in Blogger
I got a relatively easy way to highlight code snippet from http://www.craftyfella.com/2010/01/syntax-highlighting-with-blogger-engine.html.
Step-1:
Add the following code in the template just above the </head> tag.
Step-2:
Write code inside <pre> tag. For example,
The above html code gives the output as below
I tried to make it work for R highlighting, but unfortunately it is not working. So, I am using the brush for C inside the <pre> tag.
Step-1:
Add the following code in the template just above the </head> tag.
// Comment
Step-2:
Write code inside <pre> tag. For example,
// Comment
public class Testing {
public Testing() {
}
public void Method() {
/* Another Comment
on multiple lines */
int x = 9;
}
}
The above html code gives the output as below
// Comment
public class Testing {
public Testing() {
}
public void Method() {
/* Another Comment
on multiple lines */
int x = 9;
}
}
I tried to make it work for R highlighting, but unfortunately it is not working. So, I am using the brush for C inside the <pre> tag.
x <- c(1, 4, -4) m <- max(x) print(m)
ROC (Receiver Operating Characteristics) Curve in R
There are 2 packages to calculate different measures and draw different graphs of ROC cureve.
Using pROC:

- ROCR (http://rocr.bioinf.mpi-sb.mpg.de/)
- pROC (http://cran.r-project.org/web/packages/pROC/pROC.pdf)
Code to generate ROC curve and AUC (Area under the curve) value
Using ROCR: # generate ROC curve
pred <- prediction(scores, labels)
perf <- performance(pred, "tpr", "fpr")
plot(perf)
#calculate AUC value
aucPerf <- performance( pred, 'auc')
auc <- slot(aucPerf, "y.values")
Using pROC:
roc.obj <- roc(response=labels, predictor=scores)
plot(roc.obj)
auc(roc.obj)
Convert a dataframe to a vector in R
It is not possible to convert a dataframe directly to a vector. However, a dataframe can first be converted to a matrix and then to a vector.
as.vector(as.matrix(myDataFrame))
September 12, 2013
Neuron for the Beginners
Neuron is a program to simulate neuron, as the name suggests. It is highly used in neuroscience related simulation. Here are few links for the beginners:
Official Site: http://www.neuron.yale.edu/neuron/
Tutorial: http://www.anc.ed.ac.uk/school/neuron/
Syntax Highlighting in Notepad++: http://www.neuron.yale.edu/phpbb/viewtopic.php?f=30&t=1763
Official Site: http://www.neuron.yale.edu/neuron/
Tutorial: http://www.anc.ed.ac.uk/school/neuron/
Syntax Highlighting in Notepad++: http://www.neuron.yale.edu/phpbb/viewtopic.php?f=30&t=1763
September 4, 2013
Neuron: Axon, Dendrite, and Synapse
[This blog is basically a lecture note. The lecture was given by Dr. Idan Segev, a neuroscientist from the Hebrew University of Jerusalem]
Neuron
Neuron can be conceptually viewed as an input-output electric device where dendrites are the input device and the axon is the output device.
Axon
The axon is the output electrical device of neurons. It generates and carries electrical signals called "spikes".
Glial Cell: Surrounds the axon.
Dendrite
There are various types of dendrites -Purkinje Cell, Starburst amacrine cell, CA1 Pyramidal cell, etc.
Some dendrites are spiny. Dendritic spines are the regions where synapses are made on to. Typically, there are 10,000 synapse connection in a dendritic tree.
Neuron Types
Neuron can be classified based on different categories. Principal neurons project to other brain regions, while interneurons project to local regions. Usually, principal neurons are excitatory and the interneurons are inhibitatory.
Synapse
Axon and dendritic spines are separated by synapse. The axon contains small vesicles (neurotransmitter molecules). The transfer of these neurotransmitters create voltage difference in the axon and the spine. The potential in the axon cell is all or none, which is called action potential. It is like a digital signal. The potential in the spine is an analogue one. So, the synapse can be viewed as a digital-to-analogue converter.
Summary
Figure 1 summarizes all the things.
Excitatory (Red) and Inhibitatory (Cyan) axons make connection with the dendritic tree (Blue) via synapse (green). Thus the dendritic tree gets the post-synaptic potentials from the axons. All the potentials move towards the soma where all of them are summed up. If the sum of the potentials reaches a threshold, a spike is generated that moves along the axon.
Reference: Synapses, Neurons and Brains course in Coursera by Idan Segev. (link)
Neuron
Neuron can be conceptually viewed as an input-output electric device where dendrites are the input device and the axon is the output device.
 |
| Figure 1: Neuron as an I/O Device. |
Axon
The axon is the output electrical device of neurons. It generates and carries electrical signals called "spikes".
- A single, highly branched, thin (um) process emerging from the soma.
- At the "hot" axon initial segment (AIS) the spike is initiated and then propagates along the axon.
- Covered with myelin (isolating) lipic sheath, with termittent small gaps - the nodes of Ranvier (where "hot" excitable ion channels reside)
- Decorated with frequent swelling (axonal buttons/ vericocities) where the neurotransmitter "hides" (the pre-synaptic site). An axon typically has about 5000~10,000 vericocities.
 |
| Figure 2: Typical morphology of a neuron |
Glial Cell: Surrounds the axon.
Dendrite
There are various types of dendrites -Purkinje Cell, Starburst amacrine cell, CA1 Pyramidal cell, etc.
 |
| Figure 3: Types of dendritic trees. |
Some dendrites are spiny. Dendritic spines are the regions where synapses are made on to. Typically, there are 10,000 synapse connection in a dendritic tree.
 |
| Figure 4: Dendritic Spines |
Neuron Types
Neuron can be classified based on different categories. Principal neurons project to other brain regions, while interneurons project to local regions. Usually, principal neurons are excitatory and the interneurons are inhibitatory.
 |
| Figure 5: Principal neurons and interneurons. Red: Dendritic trees, Blue: Axonal trees. |
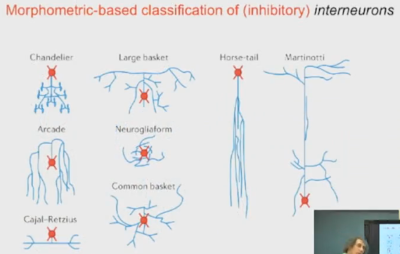 | ||
| Figure 6: Morphometric-based neuron classification |
 |
| Figure 7: Spiking pattern based neuron classification |
Synapse
 |
| Figure 8: Synapse - connecting chemical between pre-synaptic axon and post-synaptic dendritic spine. |
 |
| Figure 9: Synapse - a digital-to-analogue signal converter. |
Axon and dendritic spines are separated by synapse. The axon contains small vesicles (neurotransmitter molecules). The transfer of these neurotransmitters create voltage difference in the axon and the spine. The potential in the axon cell is all or none, which is called action potential. It is like a digital signal. The potential in the spine is an analogue one. So, the synapse can be viewed as a digital-to-analogue converter.
Summary
Figure 1 summarizes all the things.
Excitatory (Red) and Inhibitatory (Cyan) axons make connection with the dendritic tree (Blue) via synapse (green). Thus the dendritic tree gets the post-synaptic potentials from the axons. All the potentials move towards the soma where all of them are summed up. If the sum of the potentials reaches a threshold, a spike is generated that moves along the axon.
Reference: Synapses, Neurons and Brains course in Coursera by Idan Segev. (link)
July 19, 2013
July 17, 2013
How to build package in R
There is an excellent video tutorial on building package in R. The whole tutorial contains 6 short videos. I have been so impressed that I have made a playlist of my own. Here is the playlist link:
http://www.youtube.com/playlist?list=PLdvICbjqfRbdrX16ZKqq4HsKRaq4r_xSz
Enjoy R!
http://www.youtube.com/playlist?list=PLdvICbjqfRbdrX16ZKqq4HsKRaq4r_xSz
Enjoy R!
June 13, 2013
Computational Biology Problems and Challenges
Link to Few Computational Biology Problems and Challenges:
- https://genomeinterpretation.org/
- http://www.ncbi.nlm.nih.gov/books/NBK25461/#_a2000dec5ddd00344_
- http://www.youtube.com/watch?v=bVhOntMCmnQ
- http://www.frontiersin.org/Bioinformatics_and_Computational_Biology/10.3389/fgene.2011.00060/full
- http://www.stats.ox.ac.uk/research/genome/teaching2/topics_in_computational_biology
Research articles on automatic pathway construction
Research articles on automatic pathway construction:
- Inferring gene regulatory networks from gene expression data by path consistency algorithm based on conditional mutual information (2011, citation:8)
- Biological Pathway Extension Using Microarray Gene Expression Data (2008, citation:1)
- Microarray analysis of gene expression: considerations in data mining and statistical treatment (2006, citation: 66)
- Genomic analysis of metabolic pathway gene expression in mice (2005, citation: 67)
- PathMAPA: a tool for displaying gene expression and performing statistical tests on metabolic pathways at multiple levels for Arabidopsis (2003, citation: 54)
- Bayesian Consensus Pathway Construction and Expansion Using Microarray Gene Expression Data (From NCBI)
- Biological Networks (Software from UCSD)
- Reconstructing dynamic gene regulatory networks from sample-based transcriptional data (2012, citation:3)
- Reconstructing regulatory networks from the dynamic plasticity of gene expression by mutual information (2013, citation:0)
- A Gaussian graphical model for identifying significantly responsive regulatory networks from time series gene expression data (2012, citation:1)
- An integrative genomics approach to the reconstruction of gene networks in segregating populations (2004, citation:135)
- Integrating genetic and gene expression data: application to cardiovascular and metabolic traits in mice ()
- Uncovering regulatory pathways that affect hematopoietic stem cell function using 'genetical genomics' (Nature Genetics, 2005, citation:333)
- Inferring gene transcriptional modulatory relations: a genetical genomics approach (2005, citation:77)
- Complex trait analysis of gene expression uncovers polygenic and pleiotropic networks that modulate nervous system function (Nature Genetics, 2005, citation:515)
- Integrated transcriptional profiling and linkage analysis for identification of genes underlying disease (Nature Genetics, 2005, citation:410)
- An integrative genomics approach to infer causal associations between gene expression and disease (Nature Genetics, 2005, citation:544)
May 13, 2013
April 24, 2013
Latex tips and tricks
Write multiple lines in a cell of a table.
The easiest way is to use \shortstack. Few other options are suggested here.
The easiest way is to use \shortstack. Few other options are suggested here.
April 19, 2013
How to write algorithm using Latex
I got a nice introductory video on writing algorithm using Latex.
This video uses the algorithmic package. You can find the basic set of commands of this package and few alternative packages here. The detail document is available here. The algorithmicx package (link) is an advanced package for writing algorithm.
A sample code snippet for the simple bubble sort algorithm using algorithmic package and its output is given below:
\begin{algorithm}
\begin{algorithmic}
\STATE $S$ is an array of integer
\FOR {$i$ in $1:length(S)-1)$}
\FOR {$j$ in $(i+1):length(S)$ }
\IF {$S[i]> S[j]$}
\STATE swap $S[i]$ and $S[j]$
\ENDIF
\ENDFOR
\ENDFOR
\end{algorithmic}
\caption{Bubble sort algorithm}
\label{algo:bubble_sort}
\end{algorithm}
This video uses the algorithmic package. You can find the basic set of commands of this package and few alternative packages here. The detail document is available here. The algorithmicx package (link) is an advanced package for writing algorithm.
A sample code snippet for the simple bubble sort algorithm using algorithmic package and its output is given below:
\begin{algorithm}
\begin{algorithmic}
\STATE $S$ is an array of integer
\FOR {$i$ in $1:length(S)-1)$}
\FOR {$j$ in $(i+1):length(S)$ }
\IF {$S[i]> S[j]$}
\STATE swap $S[i]$ and $S[j]$
\ENDIF
\ENDFOR
\ENDFOR
\end{algorithmic}
\caption{Bubble sort algorithm}
\label{algo:bubble_sort}
\end{algorithm}
March 28, 2013
Clear all memory in R
There are several steps to clear all memory in R.
Step-1: Clear all variables from the workspace.
rm(list=ls(all=TRUE))
Note: if all=TRUE option ls function ensures to get all variables (even imported by other packages).
Step-2: Call garbage collector.
Step-1: Clear all variables from the workspace.
rm(list=ls(all=TRUE))
Note: if all=TRUE option ls function ensures to get all variables (even imported by other packages).
Step-2: Call garbage collector.
gc()
March 24, 2013
Bioinformatics Conferences and Journals
Top 10 Bioinformatics Conferences
Top 10 Bioinformatics Journals
- ISMB - Intelligent Systems in Molecular Biology
- RECOMB - Research in Computational Molecular Biology
- ECCB - European Conference on Computational Biology
- Pacific Symposium on Biocomputing
- BMEI - International Conference on BioMedical Engineering and Informatics
- CBMS - IEEE Symposium on Computer-Based Medical Systems
- WABI - Algorithms in Bioinformatics
- CMSB - Computational Methods in Systems Biology
- ICNSC - International Conference on Networking, Sensing and Control
- BIBE - IEEE International Conference on Bioinformatics and Bioengineering
Top 10 Bioinformatics Journals
- BIOINFORMATICS - Bioinformatics/computer Applications in The Biosciences
- PLOS COMPUT BIOL - PLOS Computational Biology
- BMC Bioinformatics
- BIB - Briefings in Bioinformatics
- TITB - IEEE Transactions on Information Technology in Biomedicine
- TCBB - IEEE/ACM Transactions on Computational Biology and Bioinformatics
- JBI - Journal of Biomedical Informatics
- BIOL DIRECT - Biology Direct
- JCB - Journal of Computational Biology
- CMPB - Computer Methods and Programs in Biomedicine
March 22, 2013
R subsetting may convert a dataframe into a vector
The common way of subsetting in R is to give the row or column indexes inside brackets ([rows,cols]). One notable property of this type of
However, users generally
However, users generally
Protein folding, RMSD and Gibbs Energy
Today I was discussing with my friend Swakkhar Shatabda on the protein folding problem. Although I came to know about protein folding in one of my early courses, there is a lot more to explore.
Today, I got familiar with few metrics used to measure the distance between two conformations of protein - RMSD (Root Mean Square Deviation), cRMSD and dRMSD. One ppt file helped me understand it. I got this file from Standord University website.
I also learned about Gibbs Free Energy.
I really enjoyed the discussion between us!
Today, I got familiar with few metrics used to measure the distance between two conformations of protein - RMSD (Root Mean Square Deviation), cRMSD and dRMSD. One ppt file helped me understand it. I got this file from Standord University website.
I also learned about Gibbs Free Energy.
I really enjoyed the discussion between us!
March 20, 2013
Install R in ubuntu from command line
Steps to install R in ubuntu from command line:
NOTE:
1) Step-1 is dependent on the version of ubuntu. You can find the ubuntu version using the following command:
lsb_release -a
2) You can also different mirror site for step-1. The list is available here.
3) You may need to use secure apt. In that case, just after step-1 (before step-2), run the following commands:
Reference: http://cran.r-project.org/bin/linux/ubuntu/README
- add the following configuration in /etc/apt/sources.list file
# configuration for R
deb http://cran.nexr.com/bin/linux/ubuntu precise/ - Command: sudo apt-get update
- Command: sudo apt-get install r-base
- Command to start R: R
- Command to quit R: q()
NOTE:
1) Step-1 is dependent on the version of ubuntu. You can find the ubuntu version using the following command:
lsb_release -a
2) You can also different mirror site for step-1. The list is available here.
3) You may need to use secure apt. In that case, just after step-1 (before step-2), run the following commands:
gpg --keyserver hkp://keyserver.ubuntu.com:80 --recv-keys E084DAB9
gpg -a --export E084DAB9 | sudo apt-key add -
Reference: http://cran.r-project.org/bin/linux/ubuntu/README
Continuous Query
Today I came to know a new concept in DB - Continuous Query. Traditionally, one query is run once over the current dataset and it is completed. Continuous query, in contrast, logically runs continuously over the database. Possible application of this type of query may be in Stock market, Traffic monitor etc.
Reference:
Shivnath Babu, Jennifer Widom, Continuous queries over data streams. http://ilpubs.stanford.edu:8090/527/1/2001-9.pdf
Reference:
Shivnath Babu, Jennifer Widom, Continuous queries over data streams. http://ilpubs.stanford.edu:8090/527/1/2001-9.pdf
Subscribe to:
Comments (Atom)
